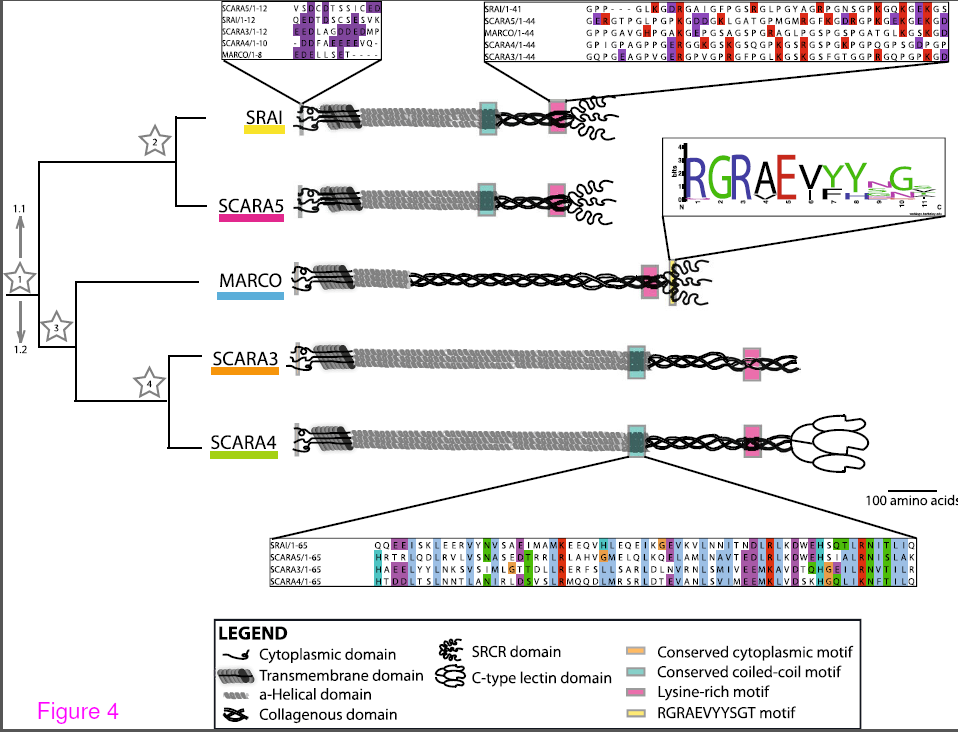
Our research on the scavenger receptor MARCO was featured in an article “Air Pollution, Evolution, and the Fate of Billions of Humans” by Carl Zimmer in the New York Times. In this manuscript we collaborated with Dr. Brian Golding, an expert in evolutionary biology in order to understand the evolution of this macrophage receptor. MARCO (or macrophage receptor with collagenous structure) is expressed on macrophages where it binds bacteria and particles such as those found in dust and air pollution. We had hypothesized that because it is the receptor for two pathogens, Streptococcus pneumoniae and Mycobacterium tuberculosis, that have played a major part in driving human evolution, that we might find evidence of areas of the receptor that were undergoing rapid evolution to protect us from this pathogen.
In order to determine which regions of the protein were changing we performed a phylogenetic analysis of the sequence of MARCO from humans, our close ancestors, the Denisovians and Neanderthals, and primates. We found a few interesting things. There was one mutation, which we call F282S (282 refers to the 282nd amino acid in the protein, the F = phenylalanine and the S= serine), had changed very rapidly. All our primate, Denisovian and Neanderthal relatives had a serine residue in that position but fully 83% of the human genomes we analyzed had a phenylalanine. The fact that this mutation spread so quickly through the population means that there must have been very strong selection pressure. We cloned both variants and found that the human specific variant was indeed better at binding inert particles and bacteria. There were a few other interesting mutations we characterized (see article below) but the take home message is that some of the evolutionary adaptations we have made to deal with pathogens may have influenced our ability to handle air pollution or, since the savannah was predicted to be a dry and dusty place, the adaptations we’ve made to deal with particulates in the air may have changed our response to pathogens.
To read the full article, see below.